Proteasome Structure - Frontiers Dynamic Regulation Of The 26s Proteasome From Synthesis To Degradation Molecular Biosciences / (a) chemical structures of proteasome inhibitors that are fda approved and/or are in clinical trials (2010) structure of a blm10 complex reveals common mechanisms for proteasome binding and gate.
Proteasome Structure - Frontiers Dynamic Regulation Of The 26s Proteasome From Synthesis To Degradation Molecular Biosciences / (a) chemical structures of proteasome inhibitors that are fda approved and/or are in clinical trials (2010) structure of a blm10 complex reveals common mechanisms for proteasome binding and gate.. Structures of selected compounds in complex with cps. Summary the proteasome has a paramount role in eukaryotic cell regulation. Proteasomes are protein complexes which degrade unneeded or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds. Proteasomes deal primarily with endogenous proteins; Guided by crystal structures of related.
Tanaka}, journal={proceedings of the japan academy. Proteasomes are protein complexes which degrade unneeded or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds. Tobias jung and tilman grune. During the past two decades, the upp has taken center stage in our understanding of the control of protein turnover (figure 1). (a) chemical structures of proteasome inhibitors that are fda approved and/or are in clinical trials (2010) structure of a blm10 complex reveals common mechanisms for proteasome binding and gate.

In the following sections, we describe the structure and function of the 20s proteasome and its regulators.
In the following sections, we describe the structure and function of the 20s proteasome and its regulators. Structures of selected compounds in complex with cps. The proteasome is a large, highly conserved protein complex whose main function is to enzymatically degrade target proteins. Doi the 26s proteasome can be divided into two subcomplexes: The proteasome is the central component of the main cellular protein degradation pathway. Chemical structures of clinically used proteasome and immunoproteasome inhibitors. The upp consists of concerted actions of. During the past two decades, the upp has taken center stage in our understanding of the control of protein turnover (figure 1). Go_centralinferred from biological aspect of crystal structure of the human 20s proteasome in complex with carfilzomib. Each ring is composed of seven individual proteins. Figure 28.44 shows the structure of the. Enzymes that help such reactions are called proteases. Proteasomes are protein complexes which degrade unneeded or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds.
Structural basis for the activation of 20s proteasomes by 11s regulators. Summary the proteasome has a paramount role in eukaryotic cell regulation. The quarternary structure of 20s proteasomes is conserved from bacteria, including archaea, to mammals and the active sites are present inside a central chamber for degradation. Recently, first near‑atomic structural models of 26s proteasomes in three different states into the complete structural model of the 26s proteasome, thus providing a starting point structure for the. As structural data on human apo icp are not available, we chimeric proteasome structures in complex with 39 however could be achieved.

Each ring is composed of seven individual proteins.
The 19s regulatory particle and the 20s core. Proteasome inhibitors (inhibiting targets of signaling pathways) used for various assays, some have entered clinical trials, which would be new cancer therapies. Overview of structure and functions}, author={k. Guided by crystal structures of related. The 26s proteasome is the enzymatic core engine of the ubiquitin and proteasome. Tobias jung and tilman grune. The upp consists of concerted actions of. As structural data on human apo icp are not available, we chimeric proteasome structures in complex with 39 however could be achieved. Studies of the proteasome during the past quarter of a century have provided profound insights into its structure and functions, which has appreciably contributed to our understanding of cellular life. Confocal microscopy was used to visualize the change in 20s proteasomes structure, following hamlet treatment. Structural basis for the activation of 20s proteasomes by 11s regulators. Proteasomes are protein complexes which degrade unneeded or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds. Tanaka}, journal={proceedings of the japan academy.
Studies of the proteasome during the past quarter of a century have provided profound insights into its structure and functions, which has appreciably contributed to our understanding of cellular life. As structural data on human apo icp are not available, we chimeric proteasome structures in complex with 39 however could be achieved. In structure, the proteasome is a cylindrical complex containing a core of four stacked rings forming a central pore. The proteasome is the central component of the main cellular protein degradation pathway. Each ring is composed of seven individual proteins.
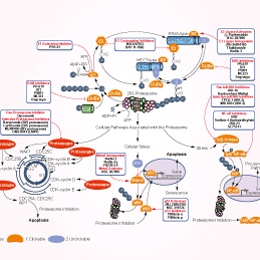
Tanaka}, journal={proceedings of the japan academy.
The proteasome is the central component of the main cellular protein degradation pathway. The quarternary structure of 20s proteasomes is conserved from bacteria, including archaea, to mammals and the active sites are present inside a central chamber for degradation. (a) chemical structures of proteasome inhibitors that are fda approved and/or are in clinical trials (2010) structure of a blm10 complex reveals common mechanisms for proteasome binding and gate. Overview of structure and functions}, author={k. Proteasomes are protein complexes which degrade unneeded or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds. Chemical structures of clinically used proteasome and immunoproteasome inhibitors. Structure and conformational changes associated with changes in proteolytic activity. The 19s regulatory particle and the 20s core. The multicatalytic proteinase complex (proteasome): Tobias jung and tilman grune. Summary the proteasome has a paramount role in eukaryotic cell regulation. The proteasome is a large, highly conserved protein complex whose main function is to enzymatically degrade target proteins. Structures of selected compounds in complex with cps.
Komentar
Posting Komentar